I tried Shopify’s TangleML as an experiment to see if I could turn my existing DCGAN notebook into a clean, reusable pipeline with caching and drop-in dataset swaps. I split the workflow into three components (prepare dataset, train GAN, generate report) so the core GAN logic stays unchanged while I can swap datasets or tune hyperparameters as separate nodes. Along the way I learned why Tangle artifacts often live as a generic data blob (portable + type-driven), and I made a tiny UX improvement by emitting a report_dir artifact that contains an index.html linking to loss_plot.png, grid.png, and downloads, so runs become viewable like a mini report page instead of hunting files manually.
I was not sure I was going in the right direction with this, or whether my implementation was correct or wrong. I just wanted to try Tangle and I already had a GAN implementation, so I thought this might be a good way. My understanding of Tangle is that it allows you to define components which in turn let you build faster. Here I kept the core GAN training principle in one component and ran different datasets through another node without changing the logic. Now other people can change parameters and hyperparameters to tune runs without touching the core principle logic.
If you want to cross-check the product framing against the original sources, start here:
Shopify Engineering announcement: Tangle
Official docs home: Tangle docs
Component authoring guide: Creating components
Source code: TangleML/tangle on GitHub
Demo of how Tangle develops on notebook
As you can see I just changed the dataset name and ran each pipeline for 2 epochs, where the base GAN logic remains intact and I get a html report for both datasets, now what if I changed something in the last node? The run would be fast as Tangle already cached the datasets and the GAN node!
Why I moved from a notebook to a pipeline
A notebook is excellent for discovery, but it tends to blur boundaries:
data prep, training, and reporting live in the same stateful session
the environment is implicit (kernel state, local packages, GPU drivers)
outputs are scattered (cells, local files, random filenames)
reruns are expensive because you accidentally redo work you could have cached
Tangle pushes you to make the boundaries explicit:
each stage has declared inputs and outputs
execution is defined (usually via a container) so runs are reproducible
artifacts are tracked systematically and can be reused
This is the same basic idea behind many ML systems: modularize stages, capture provenance, and make reruns cheap.
The mental model I used for Tangle
At a high level, I treated Tangle as:
a component specification (YAML) that defines what a stage consumes and produces
an execution runtime (containerized command) that runs the stage deterministically
an artifact store that persists outputs for inspection and reuse
a UI that lets you wire components into a graph and run experiments
A compact diagram:
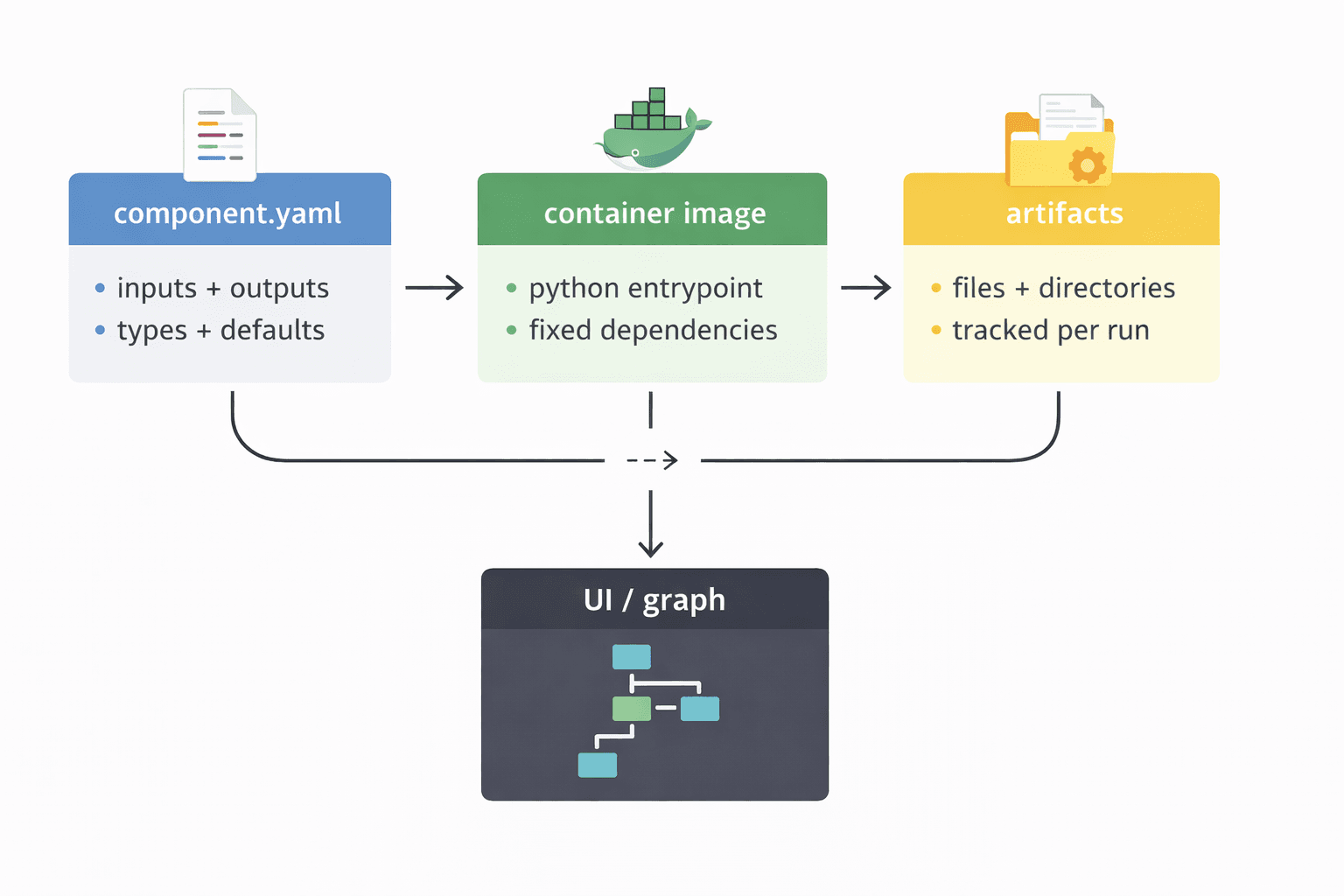
What I built
I decomposed my DCGAN notebook into three components:
prepare_tfds_images
pulls a dataset
preprocesses it into a standard artifact shape
train_dcgan
consumes the prepared dataset
trains the DCGAN
outputs a generator model and a training log
report_dcgan
consumes the generator and training log
produces plots and sample images
produces a human-viewable report bundle
Pipeline diagram:
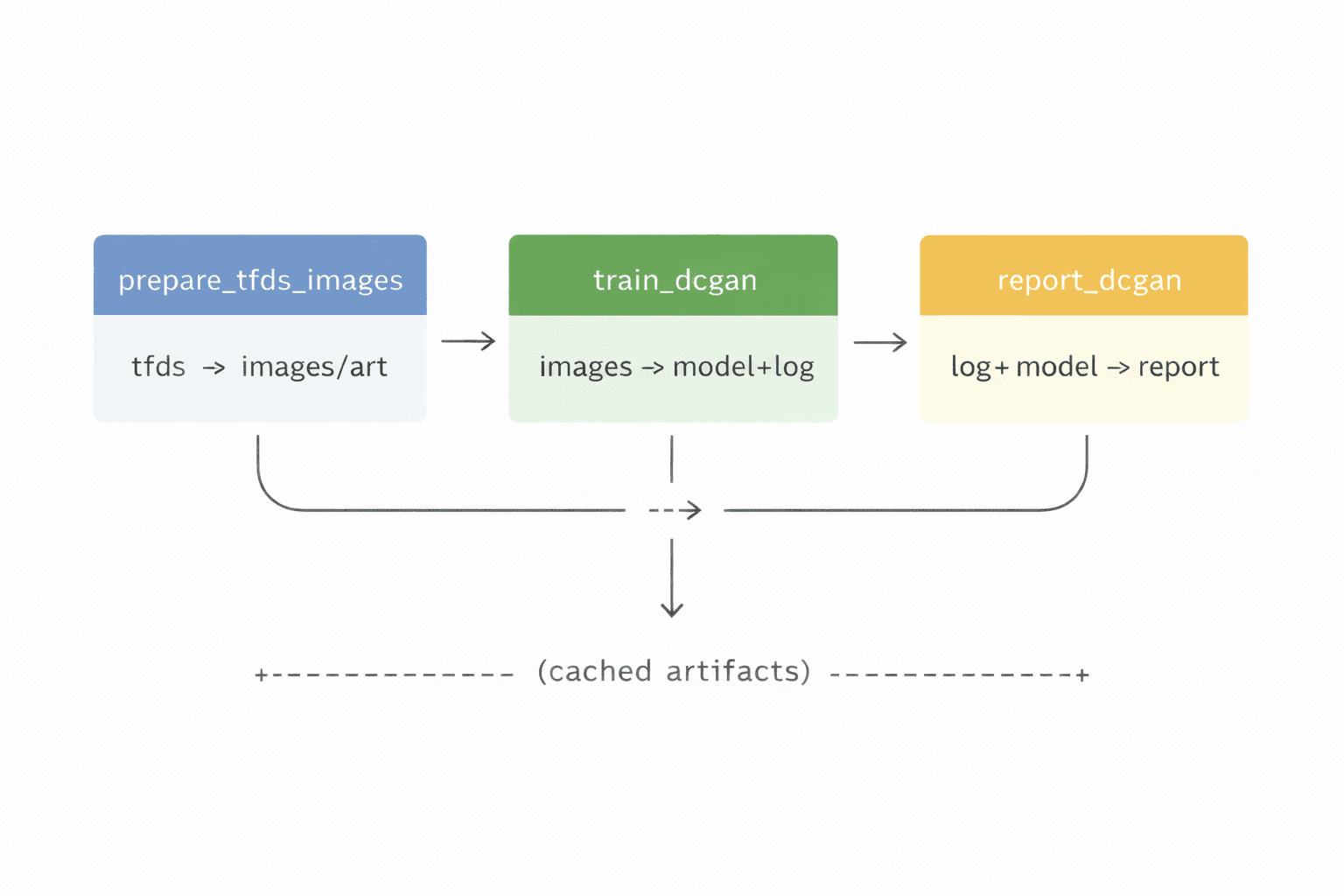
That last line is important. Once the boundaries exist, caching becomes meaningful. If I only tweak reporting, I should not retrain.
Why three YAMLs
In Tangle, the unit of reuse is the component. Each component has its own component.yaml which describes:
metadata (
name,description)typed
inputsandoutputsimplementation(how to run it, typically a container command)
So I created three YAMLs because I wanted three independent caches and three independent debug surfaces.
Practical benefit
Dataset swapping becomes a graph edit, not a code edit.
switch dataset: update inputs to prepare_tfds_images
keep GAN logic fixed: train_dcgan stays unchanged
keep reporting fixed: report_dcgan stays unchanged
How YAML, Docker, and Python are linked
Docker defines the environment
I built a single image that contains all dependencies (TensorFlow, plotting libs, TFDS, etc). Then each component points to that image.
Conceptually:
Dockerfile -> gan-tangle:latest
|
+--> report_dcgan.py
+--> train_dcgan.py
+--> prepare_tfds_images.py
YAML defines the execution contract
A key part of Tangle’s contract is the split between:
file artifacts:
{ inputPath: ... }and{ outputPath: ... }scalar params:
{ inputValue: ... }
Example pattern (from my report component):
1- "--training_log_csv"
2- { inputPath: training_log_csv }
3
4- "--latent_dim"
5- { inputValue: latent_dim }
6
7- "--out_loss_plot"
8- { outputPath: loss_plot_png }
9This distinction is what makes caching and re-execution predictable: if a scalar changes, only stages depending on that scalar should invalidate.
Python consumes paths and writes outputs
Inside the container, the Python script receives real filesystem paths and writes to them. It does not need to know anything about the UI.
A simplified mental model:
Tangle runtime
- resolves input artifacts -> mounted files/dirs
- allocates output artifact locations
- runs container command with those paths
- collects outputs back into artifact store
The artifact layout and why everything is called data
When I first inspected artifacts, this looked odd:
outputs/
loss_plot_png/
data
samples_zip/
data
run_summary_txt/
data
report_dir/
data/
grid.png
loss_plot.png
run_summary.txt
samples.npy
samples.zip
index.html
Why a file literally named data instead of loss_plot.png?
Because the artifact’s type is metadata declared in the component spec. Storing payloads at a consistent location like .../<artifact_name>/data gives a few benefits:
the storage layout is uniform for all artifact types
directories are first-class (an artifact can be a directory bundle)
tooling does not need to guess filenames or extensions
metadata and payload stay grouped by artifact name
The UX problem I ran into
The UI showed correct artifact, but the links pointed to a local filesystem path under /Users/.../data.
Example:
URI:
/Users/.../outputs/loss_plot_png/data
From the browser’s perspective, that is an HTTP URL path, not a local file path. Unless the backend explicitly serves that path, you get a 404.
So nothing was wrong with the pipeline. The missing piece was simply:
a route that maps those artifact filesystem paths to HTTP responses
plus correct headers so the browser can preview PNG and Text inline
Two ways to make artifacts human-friendly
Option A: add a minimal artifact serving route in start_local.py
This is what I did locally for best UX:
allow serving only from the artifacts root directory (safety)
sniff content type (PNG, Zip, Text)
set
Content-Disposition: inlinefor PNG and Textset
Content-Disposition: attachmentfor Zip
Request flow diagram:
Browser
GET /Users/.../outputs/loss_plot_png/data
|
v
FastAPI route
- validate path is inside data/artifacts
- return FileResponse(media_type=image/png, inline)
|
v
Browser previews image
This is a local developer experience feature. It does not change the core Tangle model, only how you view artifacts during development.
Option B: keep the pipeline outputs strict, add a report_dir
Even if you keep typed outputs for downstream pipeline connectivity (loss_plot_png, samples_zip, run_summary_txt), you can add a directory artifact:
report_diras a Directory artifactgenerate
index.htmlplus images and summaries inside it
This gives a single thing to click for “run inspection”.
In my case, I did both:
strict outputs for machines
a report bundle for humans
The HTML report improves the experience
The HTML report solves multiple issues in one place:
a single landing page for a run
stable relative paths (
loss_plot.png,grid.png, etc.)the report is portable as a directory artifact
it encourages better storytelling and debugging (plots, tables, params)
A minimal index.html (conceptual):
1<h1>DCGAN Run Report</h1>
2<img src="loss_plot.png" />
3<img src="grid.png" />
4<pre id="summary"></pre>
5<script>
6 fetch("run_summary.txt").then(r => r.text()).then(t => {
7 document.getElementById("summary").textContent = t;
8 });
9</script>
10One subtle web detail I hit: directory URLs need a trailing slash for relative asset paths to resolve the way you expect.
What this setup unlocked
Dataset swapping without touching GAN logic
I can treat the GAN training as a stable algorithm component and swap datasets upstream.
This is close to controlling variables in an experiment.
hold architecture and training loop constant
vary data distribution and hyperparameters
compare logs and samples
Caching that saves time
Once the boundaries are real, reruns become cheap:
tweak reporting, rerun only report
tweak hyperparameters, rerun training and report
tweak dataset, rerun prepare, training, report
Shareable experiments
Instead of a notebook that only works on my machine, I have:
component specs
a pinned environment (Docker image)
explicit artifacts
Which is much closer to a reproducible research artifact.
What I would improve next
If I were to push this further:
add a
metrics.jsonartifact for structured evaluationlog hyperparameters and dataset IDs into the report
include a small “run card” in the report (hash, inputs, versions)
add a simple baseline for visual quality (FID, precision/recall for GANs)
Closing thoughts
This started as a sanity check: can I take an already working GAN notebook and make it modular, reproducible, and easier to rerun on new datasets?
The answer was yes.
The only friction was artifact viewing, and that was not a modeling issue. It was a dev server UX gap. My solution was intentionally minimal: either serve artifacts safely via a small route, or treat report_dir as the human UI artifact and keep strict typed outputs for pipeline wiring.
References
github repo for entire gan without pipeline: https://github.com/Montekkundan/fashion-mnist-gan
github repo for tangle gan pipeline : https://github.com/Montekkundan/gan-tangle


